|
LAMMPS讲解76-LAMMPS实现小分子有机半导体的沉积模拟过程具体的in文件如下: ######################### ## ## ## Pentacene in file ## ## ## #########################
############################# Initial setup #################################
dimension 3 units real boundary p p f
#### Set variable ####
variable addatomscount equal count(addatoms) variable fix1count equal count(fix1) variable Aucount equal count(Au)
variable velocity equal -0.005 variable Temperature equal 350 variable D equal count(del)
############################## Potentials ###################################
atom_style full bond_style harmonic angle_style harmonic dihedral_style charmm pair_style hybrid morse 8.0 lj/cut/coul/cut 10.0 special_bonds amber timestep 1
################################ region #####################################
neighbor 2.0 bin neigh_modify delay 5 every 1 check yes
read_data Au_CH3_different_rough120.data extra/atom/types 2 extra/bond/types 2 extra/angle/types 2 extra/dihedral/types 3 extra/bond/per/atom 3 extra/angle/per/atom 7 extra/dihedral/per/atom 20 extra/special/per/atom 13
region fix1 block EDGE EDGE EDGE EDGE 0.0 3.0 units box region delete block EDGE EDGE EDGE EDGE 190.0 200.0 units box
group addatoms type 6 7 group Au type 1 group fix1 region fix1 group SAM_CH3 type 2 3 4 5 #group del dynamic all region delete every 100
################################# mass ######################################
mass 1 197 # Au mass 2 32 # S mass 3 12 # C3 mass 4 12 # C2 mass 5 1 # H
mass 6 12 # CP mass 7 1 # H
############################# LJ potential ##################################
############################## Au_UFF ###################################
pair_coeff 1 1 morse 10.954 1.583 3.024 8 pair_coeff 1 2 morse 8.763 1.470 2.650 8 pair_coeff 1 3 lj/cut/coul/cut 0.0634 3.182 pair_coeff 1 4 lj/cut/coul/cut 0.0634 3.182 pair_coeff 1 5 lj/cut/coul/cut 0.0414 2.752 pair_coeff 1 6 lj/cut/coul/cut 0.0634 3.182 pair_coeff 1 7 lj/cut/coul/cut 0.0414 2.752
############################### GAFF #####################################
pair_coeff 2 2 lj/cut/coul/cut 0.25 3.56 pair_coeff 2 3 lj/cut/coul/cut 0.15 3.48 pair_coeff 2 4 lj/cut/coul/cut 0.15 3.48 pair_coeff 2 5 lj/cut/coul/cut 0.06 3.10 pair_coeff 2 6 lj/cut/coul/cut 0.15 3.48 pair_coeff 2 7 lj/cut/coul/cut 0.06 3.10
pair_coeff 3 3 lj/cut/coul/cut 0.086 3.4 pair_coeff 3 4 lj/cut/coul/cut 0.086 3.4 pair_coeff 3 5 lj/cut/coul/cut 0.036 3.0 pair_coeff 3 6 lj/cut/coul/cut 0.086 3.4 pair_coeff 3 7 lj/cut/coul/cut 0.036 3.0
pair_coeff 4 4 lj/cut/coul/cut 0.086 3.4 pair_coeff 4 5 lj/cut/coul/cut 0.036 3.0 pair_coeff 4 6 lj/cut/coul/cut 0.086 3.4 pair_coeff 4 7 lj/cut/coul/cut 0.036 3.0
pair_coeff 5 5 lj/cut/coul/cut 0.015 2.6 pair_coeff 5 6 lj/cut/coul/cut 0.086 3.4 pair_coeff 5 7 lj/cut/coul/cut 0.086 3.4
pair_coeff 6 6 lj/cut/coul/cut 0.086 3.4 pair_coeff 6 7 lj/cut/coul/cut 0.036 3.0
pair_coeff 7 7 lj/cut/coul/cut 0.015 2.6
############################ bond coeffes ###################################
bond_coeff 1 569.4 1.3343 # C2-C2 bond_coeff 2 343.1 1.0879 # C2-H bond_coeff 3 569.4 1.3343 # C2-C3 bond_coeff 4 281.5 1.7340 # C2-S bond_coeff 5 343.1 1.0879 # C2-H
bond_coeff 6 478.4 1.387 # CP-CP bond_coeff 7 344.3 1.087 # CP-H
########################### angles coeffes ##################################
angle_coeff 1 69.3 121.81 # C2-C2-C2 angle_coeff 2 49.9 120.43 # C2-C2-H angle_coeff 3 38.1 116.90 # H-C2-H angle_coeff 4 69.3 121.81 # C2-C2-C3 angle_coeff 5 49.9 120.43 # C3-C2-H angle_coeff 6 61.2 129.37 # C2-C2-S angle_coeff 7 43.4 115.70 # S-C2-H angle_coeff 8 49.9 120.43 # C2-C3-H angle_coeff 9 38.1 116.90 # H-C3-H
angle_coeff 10 35.00 120.00 # CP-CP-H angle_coeff 11 63.00 120.00 # CP-CP-CP
########################## dihedrals coeffes ################################
dihedral_coeff 1 4 2 180 0 # C2-C2-C2-C2 dihedral_coeff 2 4 2 180 0 # C2-C2-C2-H dihedral_coeff 3 4 2 180 0 # H-C2-C2-H dihedral_coeff 4 4 2 180 0 # C2-C2-C2-C3 dihedral_coeff 5 4 2 180 0 # C3-C2-C2-H dihedral_coeff 6 4 2 180 0 # C2-C2-C2-S dihedral_coeff 7 4 2 180 0 # S-C2-C2-H dihedral_coeff 8 4 2 180 0 # C2-C2-C3-H dihedral_coeff 9 4 2 180 0 # H-C2-C3-H
dihedral_coeff 10 3.625 2 180 0 # c2-c2-c2-h dihedral_coeff 11 3.625 2 180 0 # c2-c2-c2-c2 dihedral_coeff 12 3.625 2 180 0 # h-c2-c2-h
################################# balance ###################################
comm_style tiled fix fixbalance all balance 1000 1.2 rcb
################################# thermo ####################################
minimize 1.0e-4 1.0e-6 5000 10000 thermo_style custom step temp pe etotal time thermo 1000 dump 1 all custom 10000 pentacene.xyz id type x y z
########################### initialization velocity #########################
velocity all create ${Temperature} 78273974 mom yes rot yes loop local dist gaussian
velocity fix1 set 0 0 0 units box
############################## Freeze boundary ##############################
fix f1 fix1 setforce 0.0 0.0 0.0
fix r1 all wall/reflect zhi EDGE
#### NVT ####
fix nvt1 Au nvt temp ${Temperature} ${Temperature} $(100*dt)
fix nvt2 addatoms nvt temp ${Temperature} ${Temperature} $(100*dt)
fix nvt3 SAM_CH3 nvt temp ${Temperature} ${Temperature} $(100*dt)
run 5000
################################ compute ####################################
compute Au Au temp compute SAM_CH3 SAM_CH3 temp compute add addatoms temp compute all all temp
compute_modify add dynamic/dof yes compute_modify Au dynamic/dof yes compute_modify SAM_CH3 dynamic/dof yes compute_modify all dynamic/dof yes
fix_modify nvt2 temp add dynamic/dof yes energy yes
thermo_style custom step etotal pe ke epair evdwl ecoul c_Au c_SAM_CH3 c_add c_all temp thermo 50000 thermo_modify lost ignore lost/bond ignore
################################ Deposit ####################################
molecule Pentacene Pentacene_R.txt region slab block EDGE EDGE EDGE EDGE 100 100 fix deposit_pentacene_1 addatoms deposit 800 0 250000 12345 region slab id next near 3.0 mol Pentacene vz ${velocity} ${velocity} units box
dump dump1 addatoms custom 200000 pentaceneid.dump mol id type x y z mass dump_modify dump1 sort id
################################# delete ####################################
fix d1 addatoms evaporate 10000 36 delete 49892 molecule yes
################################## Run ######################################
run 125000000
################################## nvt ######################################
write_data Au_SAM_CH3_Pentacene_ML1.data write_restart Au_SAM_CH3_Pentacene_desposit_1st.restart
print "Program successfully finish!" Pentacene分子模板如下 # pentacene molecule topology structure
36 atoms 40 bonds 66 angles 104 dihedrals
Coords
1 1.41020643 0.029404622 0.016905863 #cp 2 0.772300649 -1.209895195 0.110532533 #cp 3 1.459025378 -2.424179496 0.223790084 #cp 4 0.785318385 -3.636784401 0.317488763 #cp 5 1.513151622 -4.878998804 0.453677267 #cp 6 0.864216813 -6.051146578 0.5545002 #cp 7 -0.581490243 -6.092841482 0.522616617 #cp 8 -1.295323752 -4.938965535 0.401302087 #cp 9 -0.652762553 -3.65376428 0.302942869 #cp 10 -1.34711203 -2.480678924 0.190897082 #cp 11 -0.686946957 -1.239743872 0.089557743 #cp 12 2.341190713 0.057668601 0.025100233 #h 13 2.390043057 -2.41896526 0.235617092 #h 14 2.443796059 -4.865874741 0.472134231 #h 15 1.34864434 -6.83964628 0.64520484 #h 16 -1.024936558 -6.908790959 0.584056028 #h 17 -2.225226889 -4.983017907 0.381749096 #h 18 -2.278326812 -2.50282174 0.180295114 #h 19 -1.41020643 -0.029404622 -0.016905863 #cp 20 -0.772300649 1.209895195 -0.110532533 #cp 21 -1.459025378 2.424179496 -0.223790084 #cp 22 -0.785318385 3.636784401 -0.317488763 #cp 23 -1.513151622 4.878998804 -0.453677267 #cp 24 -0.864216813 6.051146578 -0.5545002 #cp 25 0.581490243 6.092841482 -0.522616617 #cp 26 1.295323752 4.938965535 -0.401302087 #cp 27 0.652762553 3.65376428 -0.302942869 #cp 28 1.34711203 2.480678924 -0.190897082 #cp 29 0.686946957 1.239743872 -0.089557743 #cp 30 -2.341190713 -0.057668601 -0.025100233 #h 31 -2.390043057 2.41896526 -0.235617092 #h 32 -2.443796059 4.865874741 -0.472134231 #h 33 -1.34864434 6.83964628 -0.64520484 #h 34 1.024936558 6.908790959 -0.584056028 #h 35 2.225226889 4.983017907 -0.381749096 #h 36 2.278326812 2.50282174 -0.180295114 #h
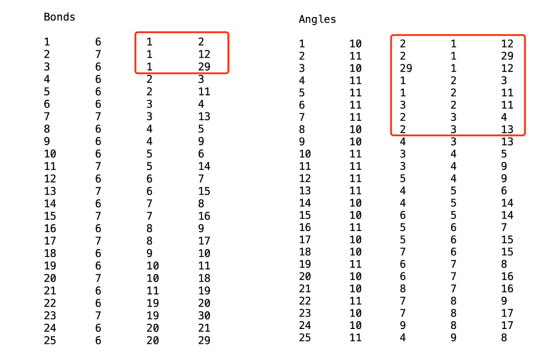
Types 1 6 2 6 3 6 4 6 5 6 6 6 7 6 8 6 9 6 10 6 11 6 12 7 13 7 14 7 15 7 16 7 17 7 18 7 19 6 20 6 21 6 22 6 23 6 24 6 25 6 26 6 27 6 28 6 29 6 30 7 31 7 32 7 33 7 34 7 35 7 36 7 Charges 1 -0.2795 2 0.1438 3 -0.2795 4 0.1438 5 -0.2795 6 -0.1011 7 -0.1011 8 -0.2795 9 0.1438 10 -0.2795 11 0.1438 12 0.15395 13 0.15395 14 0.152675 15 0.128625 16 0.128625 17 0.152675 18 0.15395 19 -0.2795 20 0.1438 21 -0.2795 22 0.1438 23 -0.2795 24 -0.1011 25 -0.1011 26 -0.2795 27 0.1438 28 -0.2795 29 0.1438 30 0.15395 31 0.153975 32 0.152675 33 0.128625 34 0.128625 35 0.152675 36 0.153975 Bonds 1 6 1 2 2 7 1 12 3 6 1 29 4 6 2 3 5 6 2 11 6 6 3 4 7 7 3 13 8 6 4 5 9 6 4 9 10 6 5 6 11 7 5 14 12 6 6 7 13 7 6 15 14 6 7 8 15 7 7 16 16 6 8 9 17 7 8 17 18 6 9 10 19 6 10 11 20 7 10 18 21 6 11 19 22 6 19 20 23 7 19 30 24 6 20 21 25 6 20 29 26 6 21 22 27 7 21 31 28 6 22 23 29 6 22 27 30 6 23 24 31 7 23 32 32 6 24 25 33 7 24 33 34 6 25 26 35 7 25 34 36 6 26 27 37 7 26 35 38 6 27 28 39 6 28 29 40 7 28 36 Angles 1 10 2 1 12 2 11 2 1 29 3 10 29 1 12 4 11 1 2 3 5 11 1 2 11 6 11 3 2 11 7 11 2 3 4 8 10 2 3 13 9 10 4 3 13 10 11 3 4 5 11 11 3 4 9 12 11 5 4 9 13 11 4 5 6 14 10 4 5 14 15 10 6 5 14 16 11 5 6 7 17 10 5 6 15 18 10 7 6 15 19 11 6 7 8 20 10 6 7 16 21 10 8 7 16 22 11 7 8 9 23 10 7 8 17 24 10 9 8 17 25 11 4 9 8 26 11 4 9 10 27 11 8 9 10 28 11 9 10 11 29 10 9 10 18 30 10 11 10 18 31 11 2 11 10 32 11 2 11 19 33 11 10 11 19 34 10 20 19 30 35 11 20 19 11 36 10 11 19 30 37 11 19 20 21 38 11 19 20 29 39 11 21 20 29 40 11 20 21 22 41 10 20 21 31 42 10 22 21 31 43 11 21 22 23 44 11 21 22 27 45 11 23 22 27 46 11 22 23 24 47 10 22 23 32 48 10 24 23 32 49 11 23 24 25 50 10 23 24 33 51 10 25 24 33 52 11 24 25 26 53 10 24 25 34 54 10 26 25 34 55 11 25 26 27 56 10 25 26 35 57 10 27 26 35 58 11 22 27 26 59 11 22 27 28 60 11 26 27 28 61 11 27 28 29 62 10 27 28 36 63 10 29 28 36 64 11 20 29 28 65 11 20 29 1 66 11 28 29 1 Dihedrals 1 10 3 2 1 12 2 10 11 2 1 12 3 11 29 1 2 3 4 11 29 1 2 11 5 11 2 1 29 20 6 11 2 1 29 28 7 10 20 29 1 12 8 10 28 29 1 12 9 11 1 2 3 4 10 10 1 2 3 13 11 11 11 2 3 4 12 10 11 2 3 13 13 11 1 2 11 10 14 11 1 2 11 19 15 11 3 2 11 10 16 11 3 2 11 19 17 11 2 3 4 5 18 11 2 3 4 9 19 10 5 4 3 13 20 10 9 4 3 13 21 11 3 4 5 6 22 10 3 4 5 14 23 11 9 4 5 6 24 10 9 4 5 14 25 11 3 4 9 8 26 11 3 4 9 10 27 11 5 4 9 8 28 11 5 4 9 10 29 11 4 5 6 7 30 10 4 5 6 15 31 10 7 6 5 14 32 12 14 5 6 15 33 11 5 6 7 8 34 10 5 6 7 16 35 10 8 7 6 15 36 12 15 6 7 16 37 11 6 7 8 9 38 10 6 7 8 17 39 10 9 8 7 16 40 12 16 7 8 17 41 11 7 8 9 4 42 11 7 8 9 10 43 10 4 9 8 17 44 10 10 9 8 17 45 11 4 9 10 11 46 10 4 9 10 18 47 11 8 9 10 11 48 10 8 9 10 18 49 11 9 10 11 2 50 11 9 10 11 19 51 10 2 11 10 18 52 10 19 11 10 18 53 11 2 11 19 20 54 10 2 11 19 30 55 11 10 11 19 20 56 10 10 11 19 30 57 10 21 20 19 30 58 10 29 20 19 30 59 11 11 19 20 21 60 11 11 19 20 29 61 11 19 20 21 22 62 10 19 20 21 31 63 11 29 20 21 22 64 10 29 20 21 31 65 11 19 20 29 28 66 11 19 20 29 1 67 11 21 20 29 28 68 11 21 20 29 1 69 11 20 21 22 23 70 11 20 21 22 27 71 10 23 22 21 31 72 10 27 22 21 31 73 11 21 22 23 24 74 10 21 22 23 32 75 11 27 22 23 24 76 10 27 22 23 32 77 11 21 22 27 26 78 11 21 22 27 28 79 11 23 22 27 26 80 11 23 22 27 28 81 11 22 23 24 25 82 10 22 23 24 33 83 10 25 24 23 32 84 12 32 23 24 33 85 11 23 24 25 26 86 10 23 24 25 34 87 10 26 25 24 33 88 12 33 24 25 34 89 11 24 25 26 27 90 10 24 25 26 35 91 10 27 26 25 34 92 12 34 25 26 35 93 11 25 26 27 22 94 11 25 26 27 28 95 10 22 27 26 35 96 10 28 27 26 35 97 11 22 27 28 29 98 10 22 27 28 36 99 11 26 27 28 29 100 10 26 27 28 36 101 11 27 28 29 20 102 11 27 28 29 1 103 10 20 29 28 36 104 10 1 29 28 36 其中值得注意的几点分别为: 1)read_data中需要注意的事项 由于我们的模拟中需要在模拟盒使用fix deposit 命令不断插入新的pentacene分子,所以在read_data命令后面需要额外注明extra/atom/…/types和extra/bond/…/per/atom,其中对于type类型的参数设置比较简单,直接找额外插入的分子中不同键角对应的type种类即可,比较难搞懂的是extra/per/atom,这里我自己摸索的理解是这样的:每个原子所具有的连接的键、角和二面角等的最大值,所以我们需要查阅插入的分子文件,如图4所示。在Bonds中,对于1号原子相关的键有三个,因此extra/bond/per/atom的值应至少大于3;在Angles中,对于2号原子相关的角有7个,因此extra/angle/per/atom的值应至少大于7,以此类推;对于extra/special/per/atom,由于我们在原有的五种原子类型的基础上额外插入了两种原子,因此在pair_coeff中与6和7号原子相关的一共有13个,因此extra/special/per/atom应至少大于13。
图4 分子模板 2)有关分子模板的制作 由于本文计算所使用的是3Mar2020版本,所以对应的分子模板也是按照此版本的手册编写的,其中为了制作沉积分子所需的单个pentacene分子模板,只需要以下几个信息即可:(1)atom、bond、angle以及dihedrals(如果有improper也需添加)的数量;(2)分子的笛卡尔坐标(Coords);(3)所有原子的类型(Types);所有原子的电荷(Charges);(4)分子中所有的键(Bonds)、角(Angles)、二面角(Dihedrals),这几项均可从单分子的data文件中获取。本文所用分子模板会放在文章最后。 3)fix deposit 命令的使用 fix deposit 命令是做分子(原子)沉积最常用的的命令,其具体的使用方法在lammps官网有很详细的说明,在这里不再赘述,我们的研究中取了一个论文中经常用到的沉积频率:每隔250000步(即0.25ns)在系统真空区域插入一个pentacene分子,一共插入了800个pentacene分子,但是由于在模拟过程中会有分子从基底脱离飞入真空层,因此我们在模拟盒真空层顶端设置一个删除区域,将脱离基底的pentacene分子迅速删除,以保证其不会影响后续分子的沉积。值得注意的是,由于在模拟过程中会不断有分子插入,因此越到模拟后期系统运行速度会下降的很厉害,对服务器的性能要求较高。
感谢鲍路瑶老师的分享,内容来自于鲍老师分享出来的资料 如有需要添加微信:lmp_zhushou 进入微信群,帮助他人,共建社区 获取完整版lammps讲义可以加微信lmp_zhushou或加入QQ群994359511 本节作者Lethe同学,感谢他的贡献
|